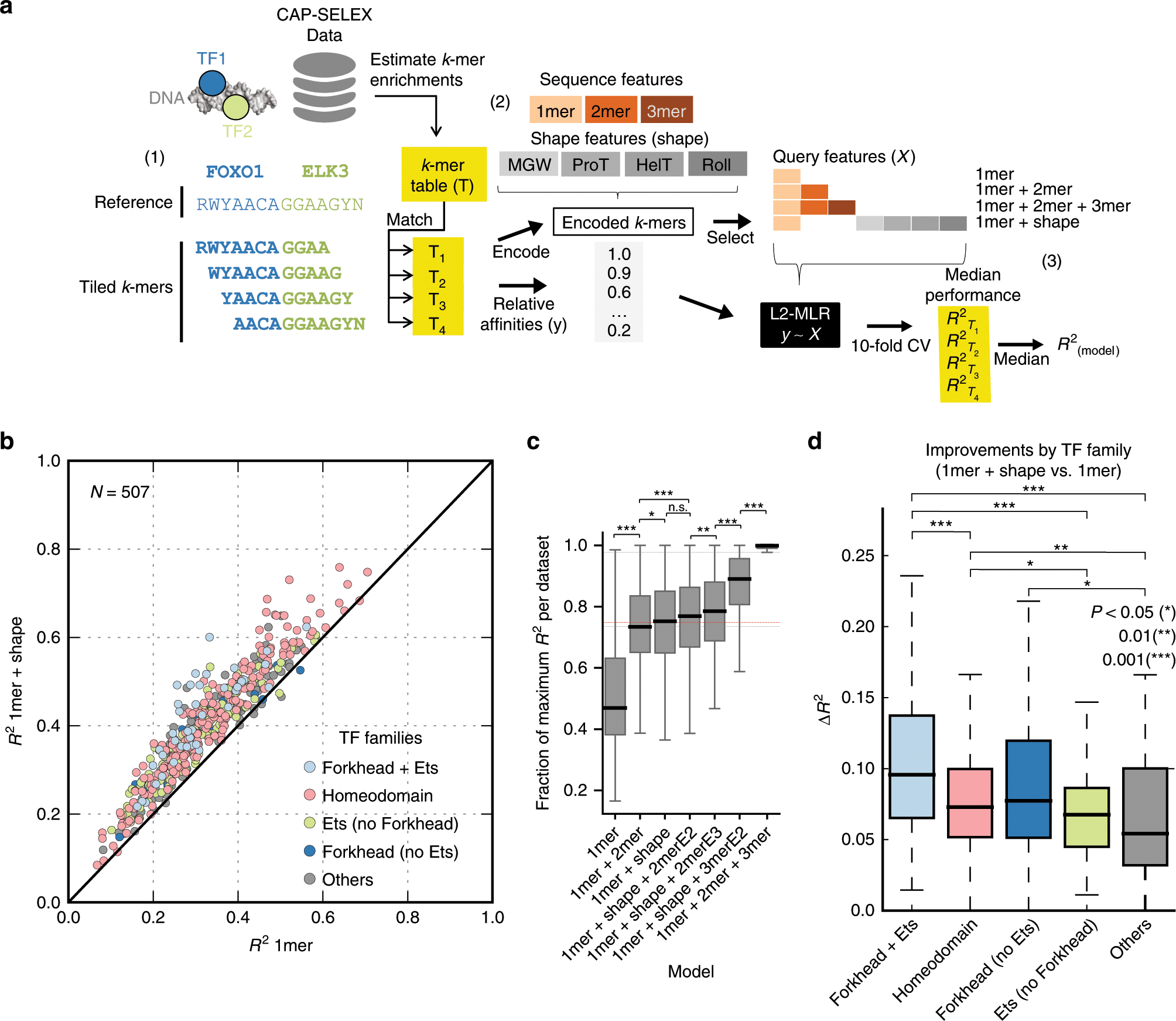
Leopard: fast decoding cell type-specific transcription factor binding landscape at single-nucleotide resolution | bioRxiv
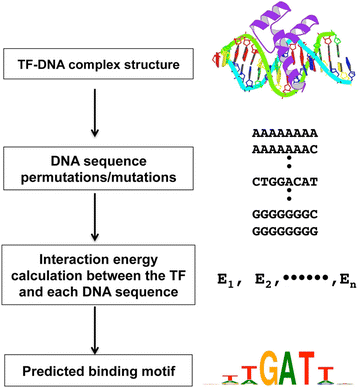
An efficient algorithm for improving structure-based prediction of transcription factor binding sites | BMC Bioinformatics | Full Text
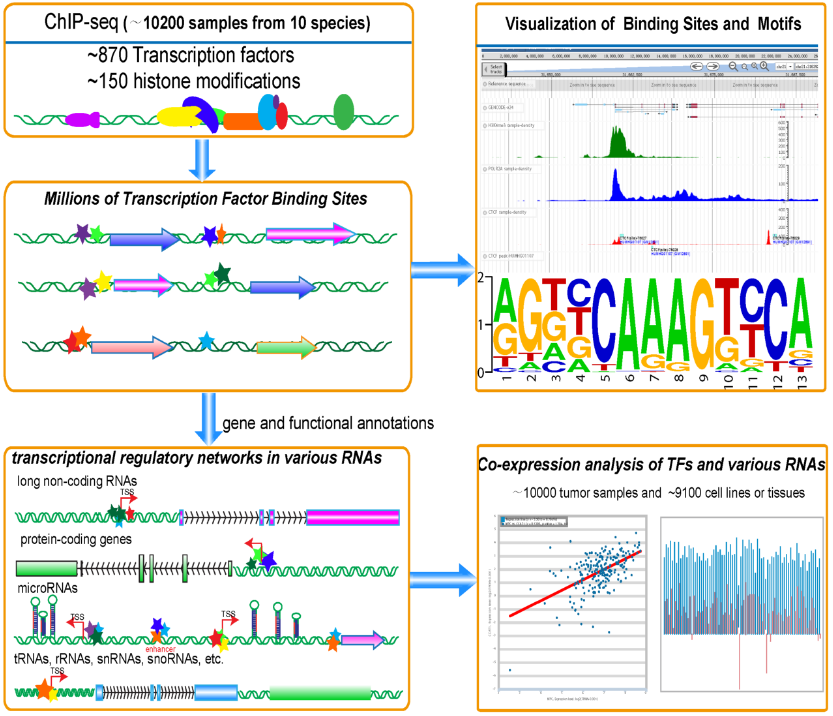
Transcription Factor Binding Sites, Motifs and Expression Profiles from ~10200 ChIP-seq and ~20000 RNA-seq samples

Prediction of transcription factor bindings sites affected by SNPs located at the osteopontin promoter - ScienceDirect

Predicting transcription factor binding sites using DNA shape features based on shared hybrid deep learning architecture: Molecular Therapy - Nucleic Acids

Prediction of transcription factor bindings sites affected by SNPs located at the osteopontin promoter - ScienceDirect
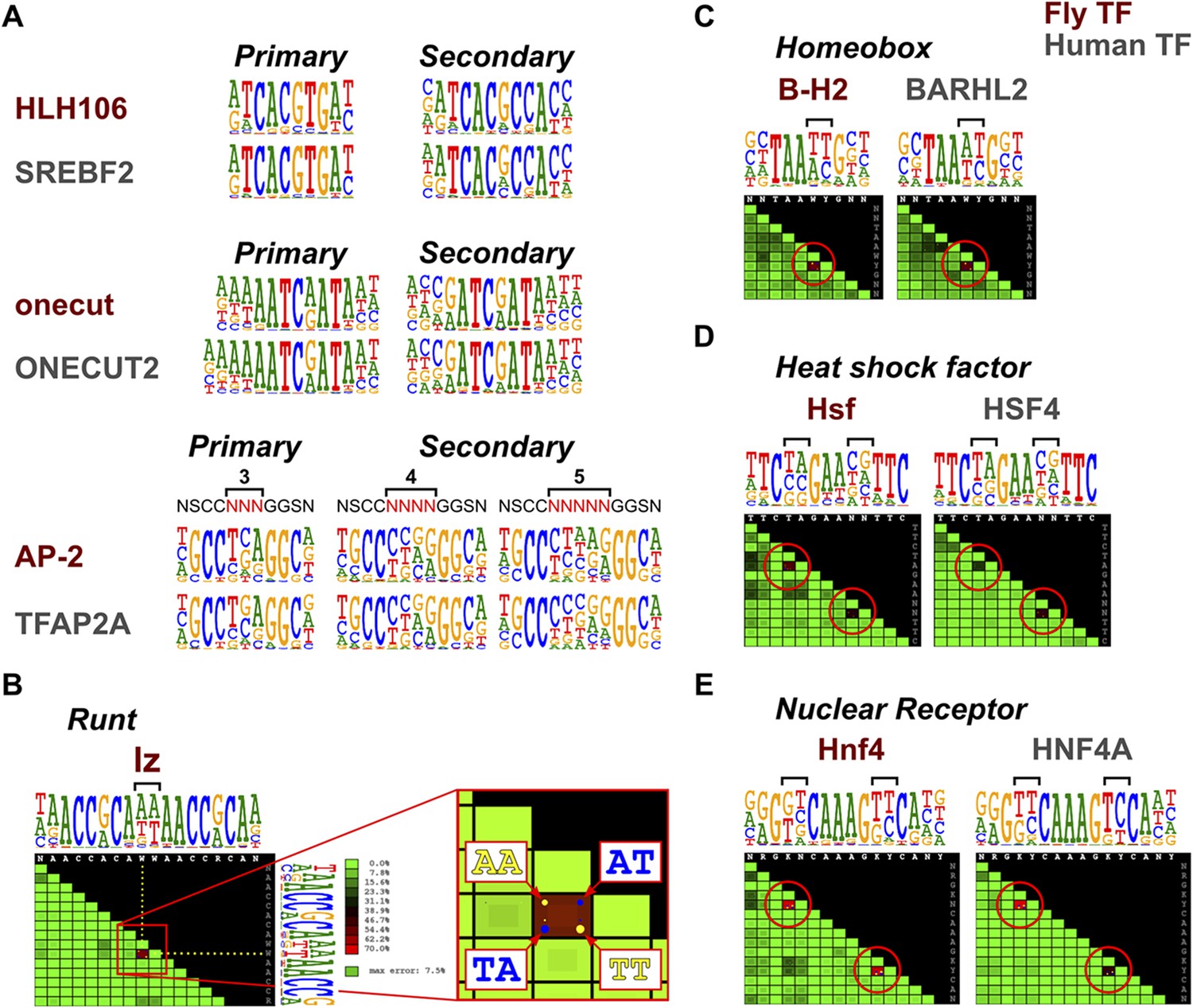
Conservation of transcription factor binding specificities across 600 million years of bilateria evolution | eLife

Prediction of transcription factor binding sites and mutation analysis... | Download Scientific Diagram

Transcriptome dynamics of developing maize leaves and genomewide prediction of cis elements and their cognate transcription factors | PNAS
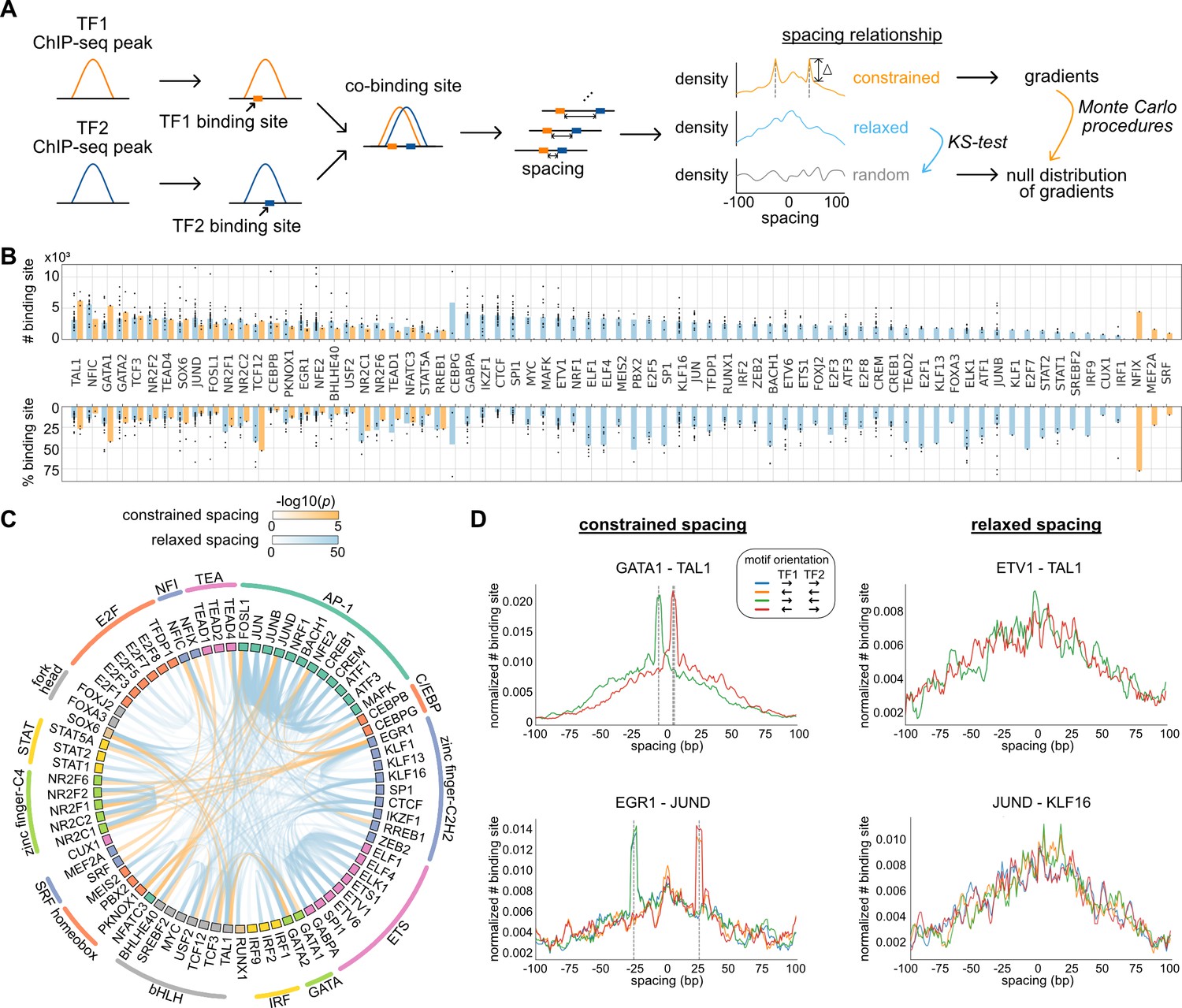
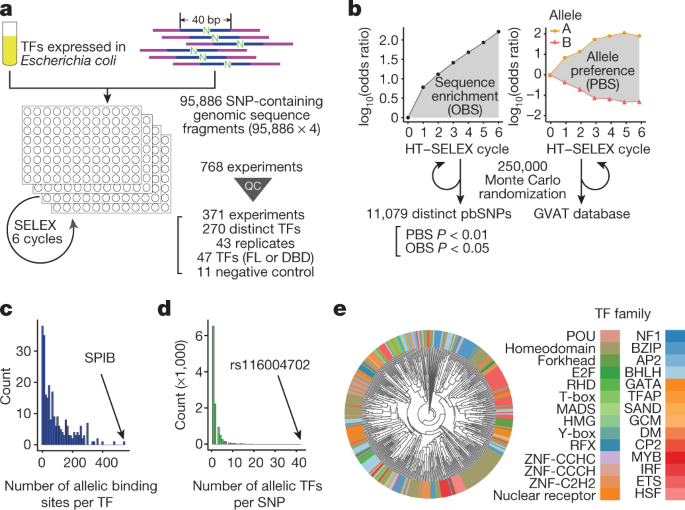
Systematic analysis of naturally occurring insertions and deletions that alter transcription factor spacing identifies tolerant and sensitive transcription factor pairs | eLife
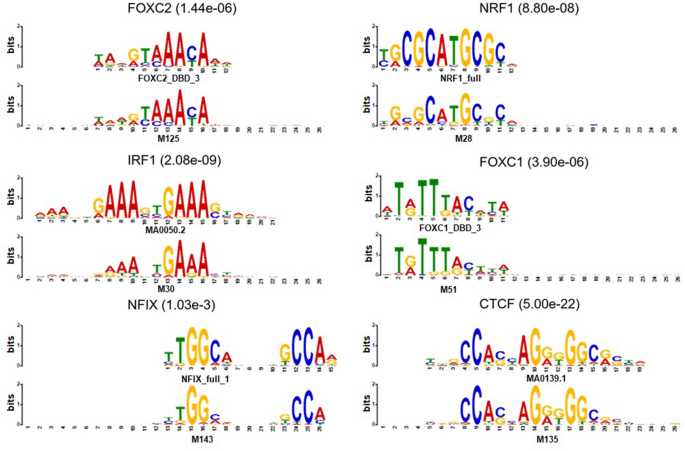
Enhancing the interpretability of transcription factor binding site prediction using attention mechanism | Scientific Reports